Inhibition of de novo lipogenesis targets androgen receptor signaling in castration-resistant prostate cancer
Giorgia Zadra,a,b,1 Caroline F. Ribeiro,a Paolo Chetta,a,c Yeung Ho,d Stefano Cacciatore,e,f Xueliang Gao,gSudeepa Syamala,a Clyde Bango,a Cornelia Photopoulos,a Ying Huang,a Svitlana Tyekucheva,h,iDebora C. Bastos,j Jeremy Tchaicha,k,2 Brian Lawney,l Takuma Uo,m Laura D’Anello,a Alfredo Csibi,k,3Radha Kalekar,a Benjamin Larimer,n Leigh Ellis,a,b,o Lisa M. Butler,p,q,r Colm Morrissey,m Karen McGovern,k,4Vito J. Palombella,k,5 Jeffery L. Kutok,k Umar Mahmood,n Silvano Bosari,s,t Julian Adams,k,6 Stephane Peluso,k,7Scott M. Dehm,d,u,v Stephen R. Plymate,m and Massimo Lodaa,b,o,1
2019 January
SIGNIFICANCE
Standard of care for metastatic castration-resistant prostate cancer (mCRPC) mainly relies on suppression of androgen receptor (AR) signaling. This approach has no lasting benefit due to the emergence of resistance mechanisms, such as ligand-independent splicing variant AR-V7. A metabolic feature of mCRPC is the upregulation of de novo lipogenesis to provide substrates and fuel for metastatic spread. Whether increased levels of fats affect AR signaling to promote an aggressive disease remains to be determined. Using a selective and potent inhibitor of fatty acid synthase we demonstrate that suppression of this key enzyme inhibits AR, most importantly AR-V7, and reduces mCRPC growth. Our findings offer a therapeutic opportunity for mCRPC and a potential mechanism to overcome resistance to AR inhibitors.
ABSTRACT
A hallmark of prostate cancer progression is dysregulation of lipid metabolism via overexpression of fatty acid synthase (FASN), a key enzyme in de novo fatty acid synthesis. Metastatic castration-resistant prostate cancer (mCRPC) develops resistance to inhibitors of androgen receptor (AR) signaling through a variety of mechanisms, including the emergence of the constitutively active AR variant V7 (AR-V7). Here, we developed an FASN inhibitor (IPI-9119) and demonstrated that selective FASN inhibition antagonizes CRPC growth through metabolic reprogramming and results in reduced protein expression and transcriptional activity of both full-length AR (AR-FL) and AR-V7. Activation of the reticulum endoplasmic stress response resulting in reduced protein synthesis was involved in IPI-9119–mediated inhibition of the AR pathway. In vivo, IPI-9119 reduced growth of AR-V7–driven CRPC xenografts and human mCRPC-derived organoids and enhanced the efficacy of enzalutamide in CRPC cells. In human mCRPC, both FASN and AR-FL were detected in 87% of metastases. AR-V7 was found in 39% of bone metastases and consistently coexpressed with FASN. In patients treated with enzalutamide and/or abiraterone FASN/AR-V7 double-positive metastases were found in 77% of cases. These findings provide a compelling rationale for the use of FASN inhibitors in mCRPCs, including those overexpressing AR-V7.
The current use of second-generation antiandrogens such as enzalutamide (Enza) or androgen synthesis inhibitors such as abiraterone (Abi) has improved quality of life and survival of patients with metastatic castration-resistant prostate cancer (mCRPC) (1–3). However, most patients eventually progress to develop treatment-refractory metastatic disease (4). Despite castrate levels of circulating androgens in patients on androgen deprivation therapy, persistent activation of the androgen receptor (AR) axis still drives mCRPC (5). Different mechanisms of resistance to AR-directed therapies have been described, including the emergence of AR splice variants (AR-Vs), such as AR-V7 (6–8). AR-V7 lacks the C-terminal ligand-binding domain of full-length AR (AR-FL) and functions as a constitutively active, ligand-independent transcription factor that drives growth of mCRPC cells in vitro and in vivo (9). AR-V7 mRNA and protein are up-regulated in mCRPC bone metastases (10) and associated with a decrease in overall survival (11) and resistance to Enza and/or Abi treatment (12).
In contrast to normal prostatic cells that rely mostly on diet-derived lipids for fatty acids (FAs), prostate cancer (PCa) progression is marked by increasing rates of de novo FA synthesis, independent of circulating lipid levels (13). The key enzyme, FA synthase (FASN), catalyzes the synthesis of palmitate from malonyl-CoA and acetyl-CoA using NADPH as the reducing agent. The expression of enzymes in the lipogenic pathway, including FASN, and transcriptional regulators such as the sterol regulatory element-binding proteins (SREBPs) is significantly increased in PCa, especially in the mCRPC setting (14–16). Besides providing structural lipids and bioenergetics to sustain cancer cell growth, enhanced FASN activity is also associated with several oncogenic mechanisms, including the activation of the PI3K/Akt/mTORC1 pathway, palmitoylation of known oncogenes including k-RAS and WNT-1, regulation of endoplasmic reticulum (ER) function to sustain membrane biogenesis (13, 17), and resistance to genotoxic insults (18). Inhibition of FASN activity through genetic and pharmacological means suppresses PCa cell growth by cell cycle arrest and/or apoptosis (13), while FASN overexpression is linked to resistance to chemotherapy (18). FASN expression is transcriptionally induced by AR through the activation of SREBP1 (19) or by a direct binding to FASN promoter regions (20). However, the evidence that SREBP1 inhibition can down-regulate AR-FL levels (21) suggests a mutual regulation between AR signaling and FA metabolism. To test this, we developed a selective and potent FASN inhibitor (IPI-9119). We used IPI-9119 to assess the metabolic and biological effects of FASN activity suppression in CRPC, including characterization of suppressive effects on AR-FL and AR-V7. Collectively, these results provide strong rationale for the design of future clinical trials with IPI-9119 in mCRPC.
RESULTS
IPI-9119 Is a Potent, Selective, and Irreversible Inhibitor of FASN. We developed an irreversible FASN inhibitor (IPI-9119) (Fig. 1A). IPI-9119 inhibits the FASN thioesterase domain by promoting acylation of the catalytic serine (SI Appendix, Fig. S1A). IPI-9119 showed high potency against human purified FASN in an in vitro biochemical assay (IC50 = 0.3 nM), with high selectivity and negligible off-target activity (SI Appendix, Fig. S1B).
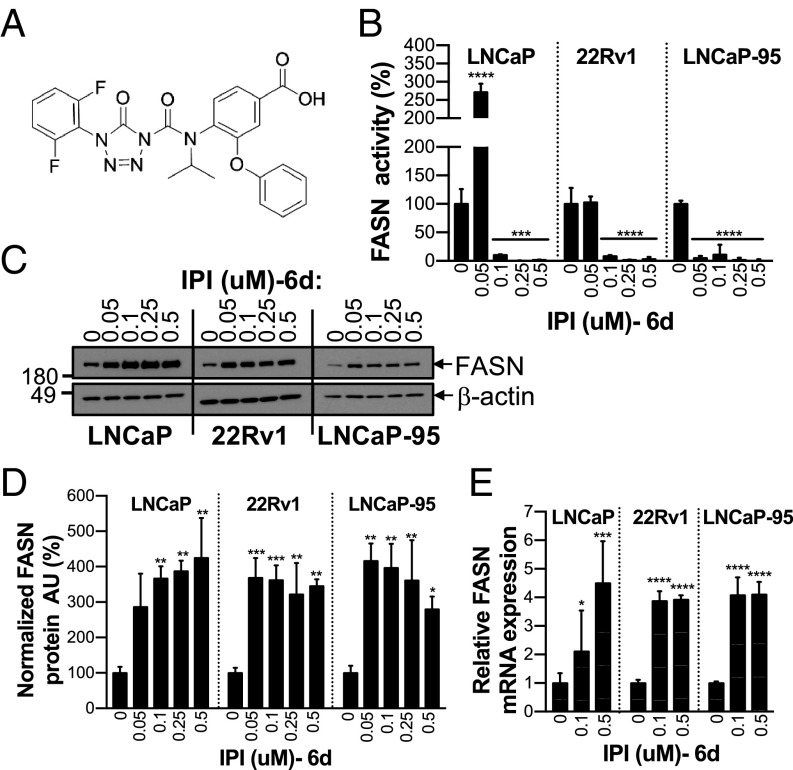
IPI-9119 is a potent and selective FASN inhibitor. (A) Chemical structure of IPI-9119 (IPI). (B) FASN activity in AD (LNCaP) and AI (22Rv1, LNCaP-95) cells, following 6-d (6d) treatment with IPI at the indicated concentrations. Data are expressed as the mean activity ± SD (n = 3) and plotted as percent DMSO. ****P < 0.0001, ***P < 0.001, one-way ANOVA followed by Tukey’s post hoc test. (C) Representative immunoblotting of FASN expression under IPI treatment. (D) Densitometric analysis. Data are expressed as normalized (FASN/β-actin) arbitrary units (AU) ± SD (n = 3) and plotted as percent DMSO. ***P < 0.001, **P < 0.01, *P < 0.05, one-way ANOVA, followed by Tukey’s post hoc test. (E) FASN mRNA levels measured by real-time qPCR. Data were normalized using β-actin as housekeeping and expressed as n-fold DMSO ± SD (n = 3 per concentration). *P < 0.05, ***P < 0.001, ****P < 0.0001, one-way ANOVA followed by Tukey’s post hoc test.
We tested FASN activity in androgen-dependent (AD) (LNCaP) and -independent (AI) (22Rv1, LNCaP-95) PCa cell lines at 3 or 6 d of incubation with IPI-9119. After 3 d of treatment, 0.05 μM IPI-9119 induced complete blockade of FASN activity. In contrast, it appeared that 0.1 μM IPI-9119 was required to sustain FASN inhibition for 6 d since FASN activity in PCa cells treated with 0.05 μM IPI-9119 was highly variable (Fig. 1B and SI Appendix, Fig. S1C). We noted increased FASN protein and mRNA expression in cells following 6 d of treatment (Fig. 1 C–E), suggesting a compensatory mechanism that required drug concentrations higher than 0.05 μM to overcome. This was also evident from RNA sequencing (RNA-seq) data of IPI-9119-treated PCa cells, which revealed broad up-regulation of lipogenic genes, as previously demonstrated with other FASN inhibitors (22, 23) (SI Appendix, Fig. S2 and Dataset S1A). Concentrations of IPI-9119 between 0.1 and 0.5 μM were selected for further investigation.
FASN Inhibition Blocks Cell Growth and Induces Apoptosis. We analyzed cell growth, cell cycle kinetics, and apoptosis across a panel of AR-positive, AD (LNCaP), AI (C4-2, C4-2B, LNCaP-Abl), and Enza/Abi-resistant AI cell lines harboring the AR-V7 splice variant (22Rv1, LNCaP-95). IPI-9119 inhibited cell growth (Fig. 2A and SI Appendix, Fig. S3A) and clonogenic survival (Fig. 2B) in PCa cell lines to a much greater extent than in the immortalized, nontransformed RWPE-1 cell line (SI Appendix, Fig. S3B). No growth inhibition by IPI-9119 was observed in FASN KO PCa cells (SI Appendix, Fig. S3 Cand D), confirming the specificity of IPI-9119 for FASN. Moreover, the antitumor effect of IPI-9119 was preserved in culture medium with whole serum compared with lipid-reduced serum (SI Appendix, Fig. S3E). Cell growth inhibition by IPI-9119 was rescued by the addition of exogenous palmitate, confirming that the effects on cell growth and survival were due to on-target activity and that palmitate rather than the toxic accumulation of malonyl-CoA accounts for the IPI-9119–mediated anticancer effect (Fig. 2C). IPI-9119 reduced the proportion of S-phase cells and increased that of G0/G1- and sub-G1–phase cells (Fig. 2D and SI Appendix, Fig. S4A) and decreased expression of cyclin A2 (Fig. 2E). Analysis of Parp cleavage, Annexin V, and propidium iodide staining showed induction of apoptosis by IPI-9119 (Fig. 2Eand SI Appendix, Fig. S4 B and C).
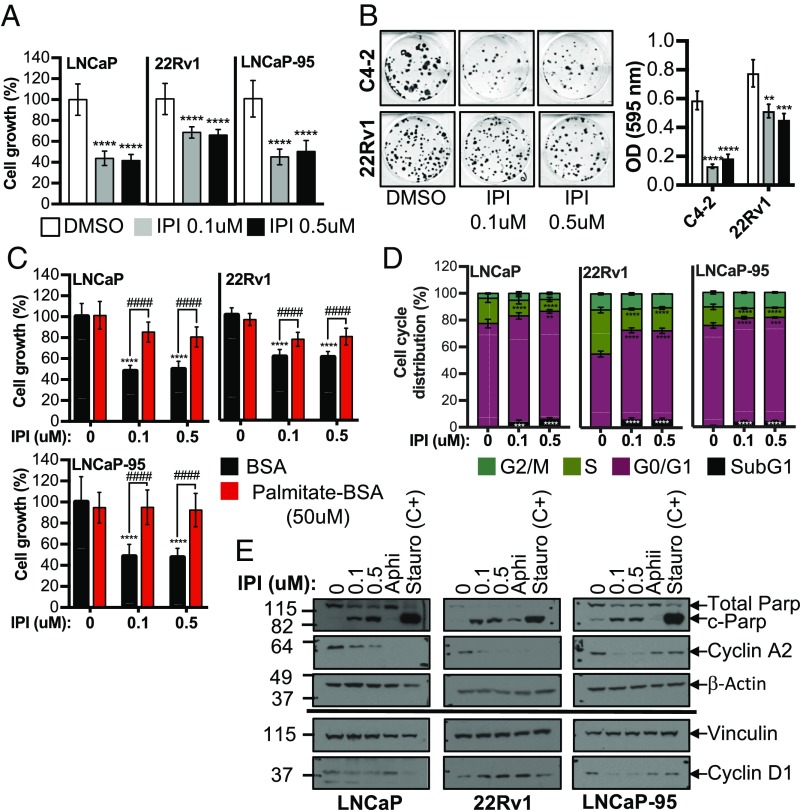
IPI-9119 inhibits PCa cell growth and induces cell cycle arrest and apoptosis. (A) Measurement of cell growth after 6-d treatment with IPI-9119 (IPI). Data are expressed as the mean number of viable cells ± SD (n per concentration = 24 for LNCaP; 15 for 22Rv1; 12 for LNCaP-95) and plotted as percent DMSO. ****P < 0.0001, one-way ANOVA followed by Tukey’s post hoc test. (B) Clonogenic assay, following 3-wk treatment with IPI or DMSO. Representative images of colony formation (Left) and quantification (Right). Data are expressed as the mean OD ± SD (n = 3). **P < 0.01, ***P < 0.001, ****P < 0.0001, one-way ANOVA followed by Tukey’s post hoc test. (C) Cell growth rescue by exogenous palmitate. Data are expressed as the mean growth ± SD (n per concentration = 9) and plotted as percent DMSO. ****P < 0.0001 IPI vs. DMSO, ####P < 0.0001 IPI + palmitate vs. IPI, two-way ANOVA followed by Sidak’s post hoc test. (D) Flow cytometry using propidium iodide (PI) and bromodeoxyuridine (BrDU), following IPI treatment (6 d). Data are expressed as the mean percent ± SD (n = 6). **P < 0.01, ***P < 0.001, ****P < 0.0001, two-way ANOVA followed by Sidak’s post hoc test. (E) Representative immunoblotting of cell cycle and apoptosis markers. Experiment was repeated twice. The S-phase inhibitor aphidicolin (Aphi, 0.3 μg/mL, 3 h) and the apoptosis inducer staurosporine (Stauro, 1 μM for 24 h) were used as positive controls. n = number of independent samples.
FASN Inhibition Alters the PCa Metabolome. The metabolic consequences of FASN activity suppression were investigated by an unbiased, global approach using untargeted MS-based metabolomics as well as biochemical assays and lipid staining. The 14C-labeling experiments and Oil Red O staining confirmed that IPI-9119 suppressed de novo FA synthesis and neutral lipid accumulation. We also found that IPI-9119 inhibited FA oxidation (FAO) due to malonyl-CoA accumulation (measured as malonyl-CoA carnitine, discussed below), resulting in the inhibition of the carnitine palmitoyltransferase 1 enzyme and FAO suppression (Fig. 3 A–C), as previously described (24). Metabolic profiling was performed in cell lysates of LNCaP, 22Rv1, and LNCaP-95 cells exposed to IPI-9119 or DMSO for 6 d. A marked separation of samples treated with IPI-9119 from control groups (independent of the drug concentration) was observed based on the entire metabolic profile (Fig. 3D). Ninety-one of 418 metabolites were significantly modulated by IPI-9119 treatment (both 0.1 and 0.5 μM) in all of the cell lines analyzed [P < 0.05; false discovery rate (FDR) < 0.05] (SI Appendix, Fig. S5 and Dataset S1B). These included metabolites involved in lipid metabolism as well as amino acid, TCA cycle, carbohydrate, and nucleotide metabolism. Gene set enrichment analysis (GSEA) using RNA-seq data confirmed down-regulation of pathways associated with amino acid and protein translation (i.e., LNCaP cells), as well as purine and pyrimidine synthesis (i.e., 22Rv1 and LNCaP-95 cells) (Dataset S1C). As expected from the suppression of de novo lipogenesis, significant reduction of the two most common FAs derived from palmitate, namely oleic and palmitoleic acids, and accumulation of polyunsaturated FAs (docosapentaenoic acid, docosahexaenoic acid, eicosapentaenoic acid, and arachidonate) (Fig. 3E, SI Appendix, Fig. S6, and Dataset S1B) were observed. Moreover, enhanced phospholipid remodeling (e.g., decrease in phosphoethanolamine species in favor to phosphatidylcholine) was induced in LNCaP and LNCaP-95 cells (Dataset S1B). Thus, while RNA-seq data highlighted compensatory transcriptional up-regulation of FA synthesis genes/regulators by IPI-9119, the intracellular metabolic changes confirmed the suppression of FASN activity.
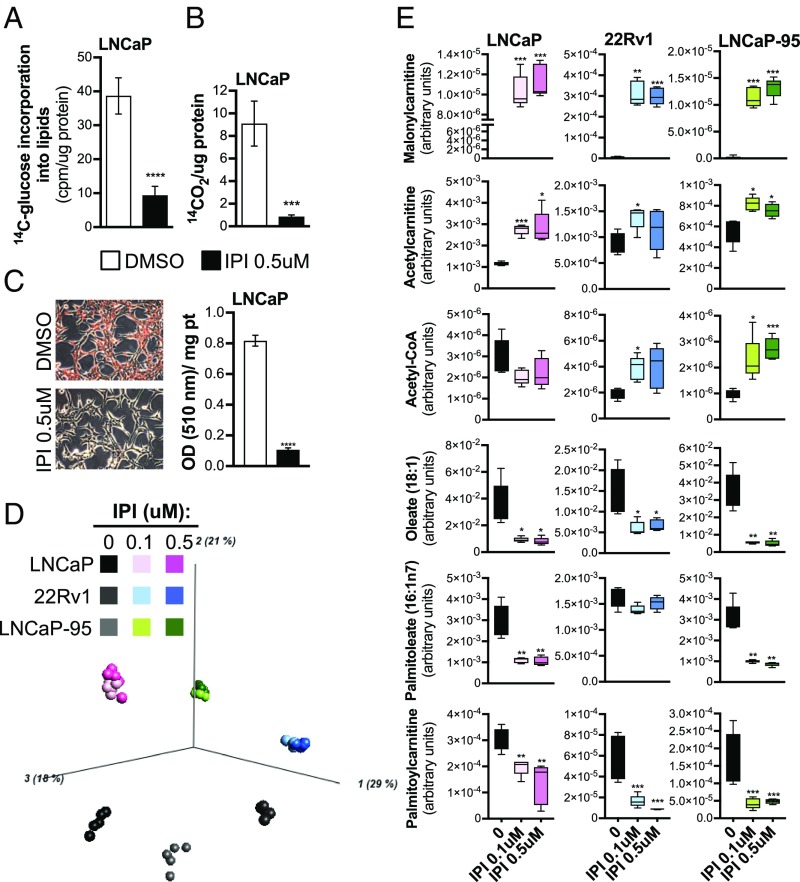
IPI-9119 induces a profound metabolic reprogramming in PCa cells. (A) Incorporation of [14C]glucose into lipids, following treatment with IPI-9119 (IPI). Results are expressed as counts per minute (cpm), normalized to protein content. Mean values ± SD are shown (n = 6); ****P < 0.0001, Student t test. (B) Fatty acid oxidation measured by 14C-CO2 release. Results are expressed as cpm normalized to protein content. Data are expressed as the mean ± SD (n = 4), normalized to protein content; ***P < 0.001, Student t test. (C) Neutral lipids accumulation in lipid droplets. Representative images (magnification: 100×) of Oil Red O staining. Quantification of neutral lipids accumulation is shown next to the images. Data are expressed as the mean absorbance ± SD (n = 3); ****P < 0.0001, Student t test. (D) Principal component analysis of the PCa cell lines analyzed (n = 6 per condition). (E) Box plots showing the levels of key metabolites involved in lipid metabolism from metabolomics data, (n = 6 per condition). ***q < 0.001, **q < 0.01, *q < 0.05, Student t test. n = number of independent samples.
Following blockade of FA synthesis, unused acetyl-CoA can be redirected toward the cholesterol pathway. Increased intracellular cholesterol levels were detected in all cell lines (SI Appendix, Fig. S7A). Accordingly, GSEA highlighted the up-regulation of cholesterol synthesis and steroidogenesis pathways (SI Appendix, Fig. S7B). MS-based analyses, however, did not reveal any significant increase in testosterone or dihydrotestosterone levels (Dataset S1D). Alterations in metabolic pathways other than FA synthesis (e.g., glutamine/glutamate, branched-chain amino acids, and glycogen metabolism) were observed in a cell-type-specific manner (SI Appendix, Fig. S6). Taken together, our data suggest that IPI-9119 profoundly alters FA metabolism, affecting both synthetic and catabolic reactions, and also impacts several related metabolic pathways.
Suppression of FA Synthesis Inhibits AR-FL and AR-V7 Expression. Since AR signaling and lipid metabolism are tightly linked, we tested the effect of the inhibition of de novo lipogenesis on AR signaling by analyzing AR expression in both AD and AI cell lines that display various patterns of AR-FL and AR-V7 expression. IPI-9119 significantly decreased AR-FL protein levels in AD LNCaP and AI C4-2 cells (expressing only AR-FL) and reduced the expression of AR-V7 in LNCaP-95 and 22Rv1 AI cells driven by this variant. Prostate-specific antigen (PSA) protein was also significantly down-regulated in AD LNCaP but not in AI cells (Fig. 4A and SI Appendix, Fig. S8A). Addition of exogenous palmitate reversed these effects, confirming that blockade of FASN activity impacts the AR/AR-V7 pathway (Fig. 4B). To rule out that AR/AR-V7 reduction was simply due to IPI-9119–mediated cell cycle arrest, treatment with the DNA replication inhibitor aphidicolin was used to mimic S phase inhibition. Despite aphidicolin-mediated complete suppression of S phase (SI Appendix, Fig. S4A), no alteration in AR/AR-V7 levels was observed (SI Appendix, Fig. S8B). These results suggest that the inhibition of AR-FL/AR-V7 protein levels is not simple reflection of IPI-9119–mediated cell cycle arrest.
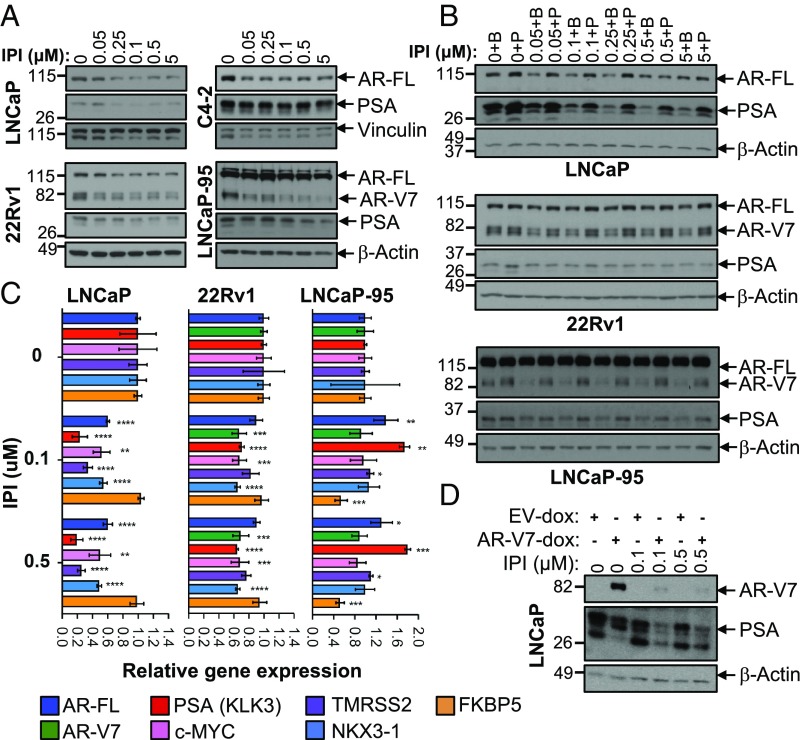
IPI-9119 inhibits AR-FL and AR-V7 protein expression. (A) Representative immunoblotting showing the reduction of AR-FL and AR-V7 in AD and AI cells. Experiment was repeated three times. (B) Representative immunoblotting showing palmitate-mediated rescue of AR-FL and AR-V7 inhibition. Palmitate (50 μM) was complexed with BSA (molar ratio 6:1), while control cells were treated with BSA only. Experiment was repeated three times. (C) Real-time qPCR of AR-FL/AR-V7 and AR-target genes. Data are plotted as the mean of three independent samples per condition and expressed as fold of DMSO (set as 1) ± SD. (D) Representative immunoblotting showing IPI-mediated ectopic inhibition of AR-V7 in negative LNCaP cells using a doxycycline-inducible AR-V7 construct. Experiment was repeated three times, using independent infections.
IPI-9119 inhibits AR-FL and AR-V7 protein expression. (A) Representative immunoblotting showing the reduction of AR-FL and AR-V7 in AD and AI cells. Experiment was repeated three times. (B) Representative immunoblotting showing palmitate-mediated rescue of AR-FL and AR-V7 inhibition. Palmitate (50 μM) was complexed with BSA (molar ratio 6:1), while control cells were treated with BSA only. Experiment was repeated three times. (C) Real-time qPCR of AR-FL/AR-V7 and AR-target genes. Data are plotted as the mean of three independent samples per condition and expressed as fold of DMSO (set as 1) ± SD. (D) Representative immunoblotting showing IPI-mediated ectopic inhibition of AR-V7 in negative LNCaP cells using a doxycycline-inducible AR-V7 construct. Experiment was repeated three times, using independent infections.
To further clarify how IPI-9119 reduces AR and AR-V7 levels, we first determined the effects of IPI-9119 on AR-FL and AR-V7 expression at the transcriptional level by real-time qPCR. While AR-FL mRNA was significantly reduced in IPI-9119-treated LNCaP, no significant effect on AR transcripts other than mild down-regulation of AR-V7 in 22Rv1 was observed in AI lines (Fig. 4C). Confirmation that AR-V7 was mainly regulated at the protein level came after administering IPI-9119 to LNCaP cells stably overexpressing AR-V7 under the inducible TRE3G promoter. Ectopic AR-V7 protein expression was significantly reduced by IPI-9119 treatment (Fig. 4D). The use of a second FASN inhibitor (TVB-3166) (SI Appendix, Fig. S8C) and FASN knockdown (KD) (SI Appendix, Fig. S8 D and E) or KO (SI Appendix, Fig. S3D) recapitulated the effect of IPI-9119 on AR-FL and AR-V7 proteins. Furthermore, the expression of nuclear AR-FL and AR-V7 was dramatically diminished by IPI-9119 in all of the PCa cell lines analyzed (SI Appendix, Fig. S9 A and B). Finally, the expression of key proteins of pathways associated with cell survival, proliferation, and energy stress (i.e., Akt/S6RP/4E-BP1, MAPK, beta-catenin, STAT3, AMPK) was analyzed to exclude that AR/AR-V7 reduction is a mere result of global cell stress response. Neither significant inactivation of these pathways nor activation of the energy sensor AMPK was observed (SI Appendix, Fig. S10). Rather, increased phosphorylation of Akt was observed in both LNCaP and LNCaP-95 cells concomitant to IPI-9119–mediated AR/AR-V7 reduction, suggesting a compensatory reciprocal feedback, as previously described (25). Only c-MYC and AR/AR-V7 proteins were consistently reduced in all of the models analyzed (Fig. 4 and SI Appendix, Fig. S10).
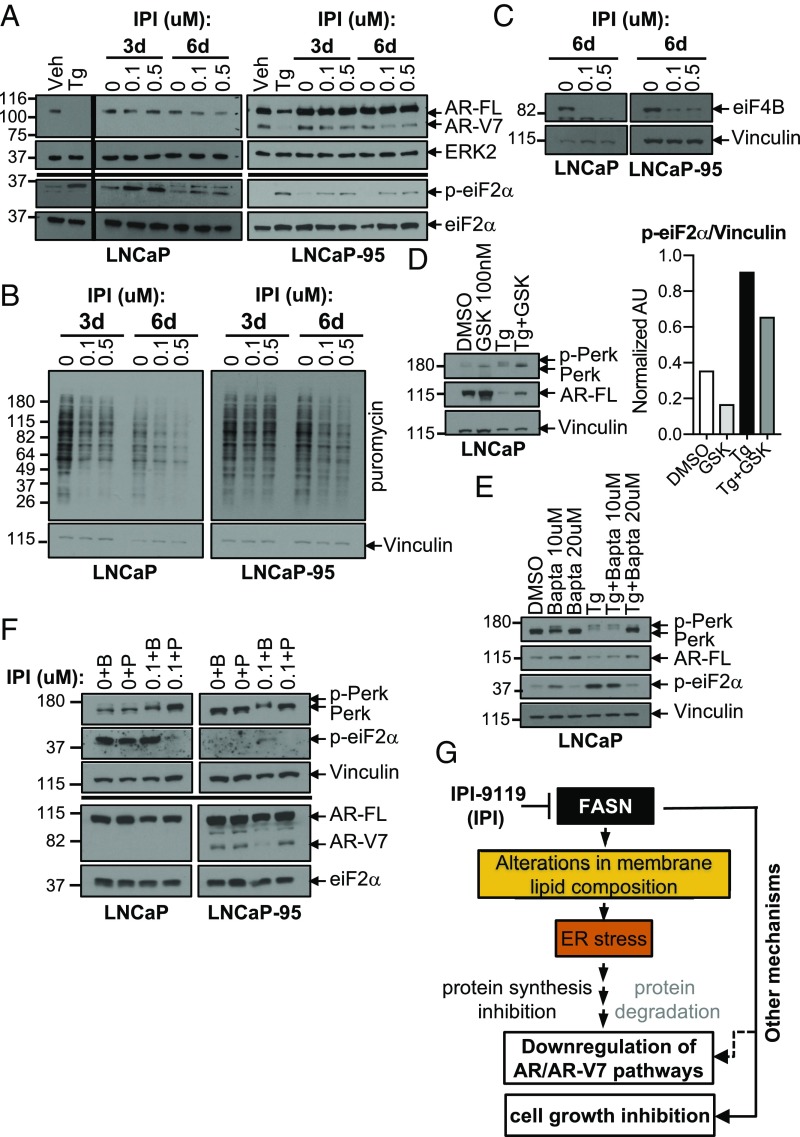
IPI-9119 Induces ER Stress and AR/AR-V7 Protein Translation Inhibition. Changes in membrane lipid composition, including increased phosphatidylcholine/phosphatidylethanolamine ratio and increased free cholesterol levels induce ER stress (26–28). Our metabolomics as well as metabolite set enrichment analyses (MSEA) using lipidomics data confirmed these alterations following IPI-9119 treatment (Dataset S1 B and E–I). FASN inhibition-induced ER stress was previously associated with increased phosphorylation of the translation initiation factor eiF2α, reduction in cellular protein synthesis, and induction of apoptosis (17). Moreover, alteration of calcium (Ca2+) homeostasis induced by the ER stress inducer thapsigargin (Tg) or Ca2+ ionophore was reported to inhibit Cap-dependent AR protein synthesis and to induce AR degradation, respectively (29, 30). Therefore, we assessed whether induction of ER stress was associated with IPI-9119–mediated reduction in AR-FL/AR-V7 protein synthesis and antiproliferative effects. We confirmed increased phosphorylation of eiF2α starting at 3 d of treatment with IPI-9119 and global protein synthesis reduction (Fig. 5 A and B). Accordingly, we observed IPI-9119–mediated reduction of protein translation cofactor eiF4B, recently involved in FASN-mediated oncogenic translation (31) (Fig. 5C). To corroborate the link between induction of ER stress and reduction of AR axis, we used the ER stressor Tg at early time points (12 h) where cell growth inhibition was not evident yet (SI Appendix, Fig. S11A). While Tg induced significant reduction of AR-FL in LNCaP cells, a 1-h pretreatment with an ER stress inhibitor, the Ca2+ chelant bapta (20 μM), partially restored its expression. Similar results were obtained when ER stress was inhibited using the PERK inhibitor GSK 2606414 (100 nM, 24 h) (Fig. 5 D and E). Accordingly, treatment with the specific inhibitor of the eiF2α phosphatase, salubrinal, induced AR/AR-V7 reduction (SI Appendix, Fig. S11B). These results suggest that ER stress is involved in inducing AR pathway down-regulation. Finally, addition of exogenous palmitate ameliorates ER stress markers while rescuing both AR/AR-V7 expression (Fig. 5F) and IPI-9119–induced cell growth inhibition (Fig. 2C). We confirmed these findings using FASN KO models (SI Appendix, Fig. S11C). Altogether, these data suggest the participation of ER stress in mediating the anticancer effect of FASN inhibition (Fig. 5G) in concert with other mechanisms, first and foremost the reduction of lipid membrane synthesis. No evident induction of autophagy was observed in association with ER stress (SI Appendix, Fig. S11D).

IPI-9119 induces ER stress response and protein translation inhibition. (A) Immunoblotting showing the induction of the ER stress marker p-eiF2α in LNCaP and LNCaP-95 cells treated with IPI-9119 (IPI) for 3 and 6 d. (B) SUnSET assay using lysates from LNCaP and LNCaP-95 cells treated with IPI for 3 and 6 d. (C) Representative immunoblotting showing the down-regulation of translation initiaton cofactor eiF4B, following treatment with IPI for 6 d. Experiment was repeated twice (LNCaP) and four times (LNCaP-95). (D) Immunoblotting showing the rescue of AR expression in LNCaP, following reduction of ER stress using the PERK inhibitor GSK2606414 (100 nM, 24 h; 1-h pretreatment). Phosphorylation of PERK is shown as an upward shift of the protein molecular weight. (E) Immunoblotting showing the rescue of AR expression in LNCaP, following reduction of ER stress using the Ca2+ chelant bapta (10, 20 μM, 12 h; 1-h pretreatment). Next to the immunoblotting, densitometric analysis of p-eiF2a is shown. Values are reported as normalized arbitrary units (AU). (F) Representative immunoblotting showing concomitant rescue of IPI-mediated AR/AR-V7 reduction and ER stress, following incubation with palmitate for 3 d. Palmitate (50 μM) was complexed with BSA (molar ratio 6:1), while control cells were treated with BSA only. (G) Schematic representation of the involvement of ER stress in mediating IPI effects.
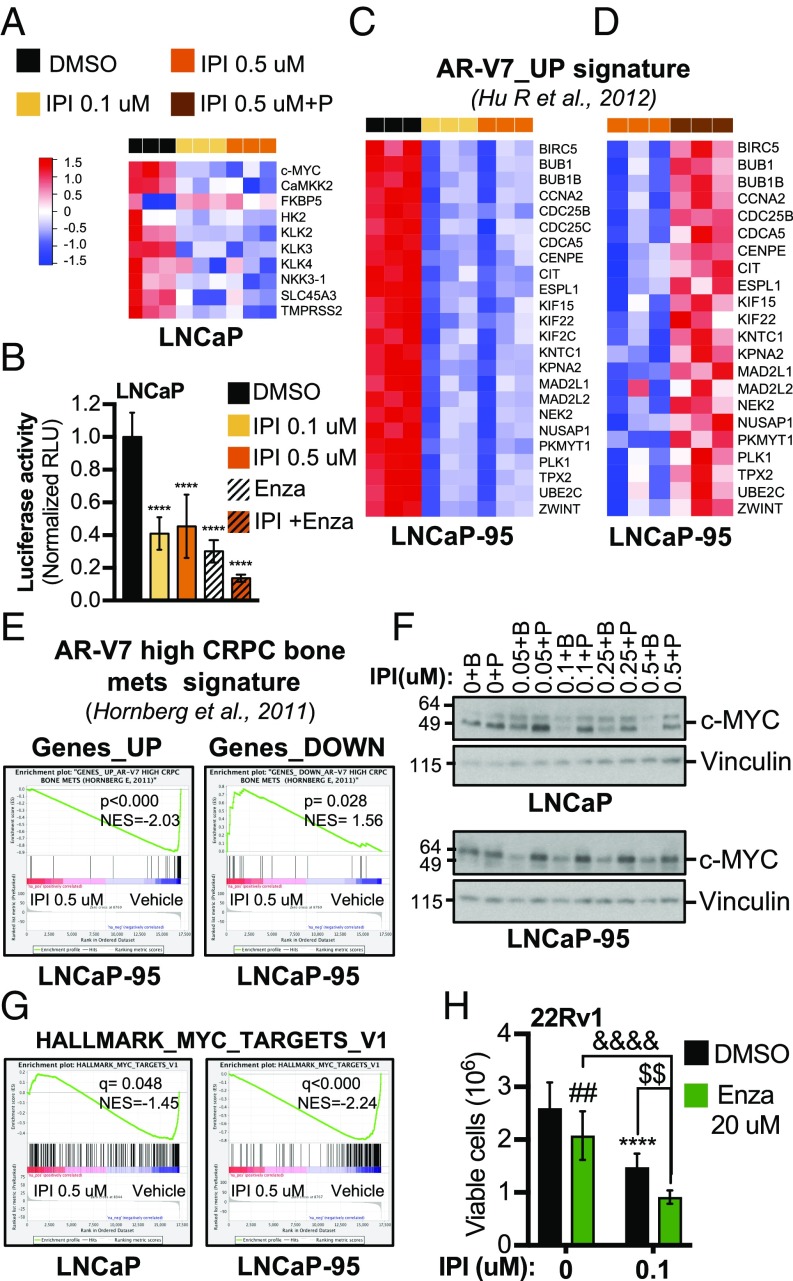
Suppression of FA Synthesis Inhibits AR-FL and AR-V7 Transcriptional Activity. Analysis of both RNA-seq and real-time qPCR data from LNCaP cells treated with IPI-9119 revealed reduced expression of canonical AR-FL target genes (e.g., KLK3, TMRSS2, and NKX3.1) (Figs. 4C and and6A).6A). Accordingly, activity of an AR-responsive luciferase reporter was inhibited by IPI-9119 in LNCaP cells (Fig. 6B). Conversely, in LNCaP-95 cells, where IPI-9119 primarily inhibits AR-V7, the expression of FKBP5 but not KLK3 (PSA) was inhibited by IPI-9119 (Fig. 4C). We observed suppression of an M-phase cell cycle gene signature previously found to be associated with AR-V7 transcriptional activity (32), also representing biphasic AR-FL targets (Fig. 6C) (7). Strikingly, this signature was completely restored by the addition of exogenous palmitate (Fig. 6D and Dataset S1L). More importantly, IPI-9119 inhibited a gene signature found in CRPC bone metastases, which express high mRNA levels of AR-V7 (33) (Fig. 6E). Similar results were obtained in 22Rv1 cells (SI Appendix, Fig. S12 A and B). Moreover, we observed trending down-regulation of a recently published AR-V7-modulated gene set (33 up-regulated genes in common between LNCaP-95 and 22Rv1 cells) identified during analyses of AR-V7–regulated transcriptome and cistrome (34) (SI Appendix, Fig. S12C). c-MYC, an oncogene commonly amplified in CRPC, had been previously described as an androgen-independent, AR-dependent target gene, which we confirmed in this study (SI Appendix, Fig. S12D). c-MYC protein was significantly reduced in IPI-9119–treated AD and AI cells and rescued by exogenous palmitate (Fig. 6F). Preranked GSEA also confirmed the down-regulation of a c-MYC transcriptional signature (V1) (Fig. 6G). Thereafter, we investigated the role of c-MYC in the response to IPI-9119 treatment. Thus, we treated c-MYC–overexpressing PCa cells derived from Hi-MYC mice (35) with IPI-9119 for 6 d. MYC-CaP cells showed significant cell growth inhibition following IPI-9119 treatment (around 40% and 70% at 0.1 and 0.5 μM, respectively). These data suggest a mutual regulation between FASN and c-MYC whereby c-MYC determines a dependency on de novo FA synthesis and FASN inhibitors shut off this key metabolic and oncogenic driver (SI Appendix, Fig. S12 E and F).
IPI-9119 inhibits AR transcriptional activity and enhances Enza efficacy. (A) Heat map of canonical AR target genes, following treatment with IPI-9119 (IPI) or DMSO for 6 d. Normalized counts (n = 3) are shown. (B) Luciferase activity in LNCaP cells treated with IPI for 6 d. ****P < 0.0001, one-way ANOVA, followed by Tukey’s post hoc test. Data represent mean ± SD (n = 3). (C) Heat map of RNA-seq data showing IPI-mediated abrogation of the AR-V7_UP gene signature after 6 d of treatment. Normalized counts are shown (n = 3). (D) Heat map of TaqMan Array Microfluidic Cards data showing palmitate rescue of the AR-V7_UP gene signature. Results are expressed as fold change of DMSO treatment. Normalized values are shown (n = 3). (E) Preranked GSEA analysis showing IPI-mediated reversion of a gene signature associated with CRPC bone metastases expressing high levels of AR-V7; P values are indicated. (F) Representative immunoblotting showing the reduction of c-MYC protein expression under treatment with IPI for 6 d. Coincubation with palmitate restored c-MYC expression. Palmitate (50 μM) was complexed with BSA (molar ratio 6:1), while control cells were treated with BSA only. (G) Preranked GSEA analysis showing IPI-mediated negative modulation of MYC_TARGETs_V1 signature (Hallmarks; h.all.v5.2s symbols.gmt); FDR values are indicated. (H) Cell growth after 6 d of Enza and IPI cotreatment. ****P < 0.0001 IPI vs. DMSO, ##P < 0.01 Enza vs. DMSO, $$P < 0.01 IPI+Enza vs. IPI, &&&&P < 0.01 IPI+Enza vs. Enza, two-way ANOVA followed by Sidak’s post hoc test. n = number of independent samples.
Finally, we went back to our findings of IPI-9119–mediated reduction of AR-V7 protein to test the combination of IPI-9119 and Enza in 22Rv1, a cell line resistant to Enza and driven by AR-V7. Our data show that the combination of IPI-9119 and Enza was more effective in reducing 22Rv1 cell growth than either of the single agents (Fig. 6H).
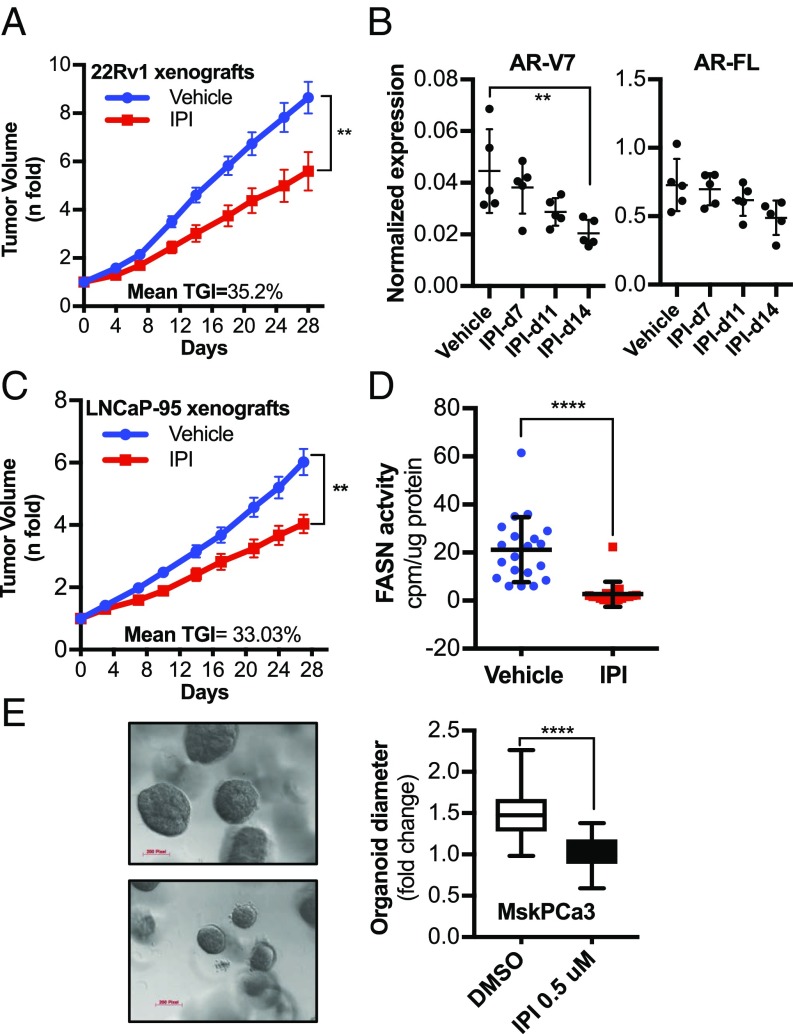
FASN Inhibition Reduces Tumor Growth in CRPC Preclinical Models and Human Organoids. To translate our in vitro findings into the preclinical setting, we investigated the effect of IPI-9119 in mouse models of CRPC resistant to Enza and/or Abi; 22Rv1 and LNCaP-95–bearing xenografts were exposed to IPI-9119 with a constant infusion (0.5 μL/h; 100 mg/mL) for 4 wk, using an s.c. osmotic ALZET pump. Mice did not show any signs of toxicity, stress, weight loss (SI Appendix, Fig. S13A), or changes in feeding behavior. We observed significant inhibition of 22Rv1 xenograft tumor growth at the end of the treatment (35% inhibition at end of treatment; P < 0.0056, Mann–Whitney U test) (Fig. 7A) and reduced AR-V7 levels following 2 wk of treatment (Fig. 7B). Similar results were obtained in mice bearing LNCaP-95 xenografts (33% inhibition at end of treatment; P < 0.0016, Mann–Whitney U test) (Fig. 7C), with concomitant intratumoral inhibition of FASN activity (Fig. 7D). In line with the cell line data, cholesterol levels and the expression of the key enzyme in cholesterol synthesis, 3-hydroxy-3-methylglutaryl-CoA reductase were increased also in vivo (SI Appendix, Fig. S13 B and C). Finally, we tested the efficacy of IPI-9119 in MSK-PCa 3, which is a FASN- and AR-FL–positive human mCRPC organoid line (36). An equal number of cells were plated and organoids were grown for 25 d. We observed significant reduction in organoid growth under treatment with IPI-9119 (Fig. 7E), confirming the antitumorigenic effect of FASN inhibition in human mCRPC.
IPI-9119 inhibits tumor growth of CRPC xenografts and human mCRPC-derived organoids. (A) Average tumor volume of 22Rv1 xenografts during 28-d treatment with IPI-9119 (IPI) using the ALZET s.c. pump infusion (n= 12 vehicle, n = 11 IPI-9119). Results are expressed as n-fold the mean initial volume (equal to 1) ± SEM. (**P = 0.0056, end of treatment, Mann–Whitney nonparametric test). (B) Densitometric analysis of AR-FL and AR-V7 protein expression in xenograft homogenates at the time points analyzed (**P = 0.0091, ANOVA test, followed by Tukey’s post hoc test). (C) Average tumor volume of LNCaP-95 xenografts (n = 20 vehicle, n= 17 IPI) treated as in A. Results are expressed as n-fold the mean initial volume (equal to 1) ± SEM (**P = 0.0016, end of treatment, Mann–Whitney nonparametric test). (D) Measurement of FASN activity in LNCaP-95 xenografts homogenates collected at the end of treatment. Results are expressed as counts per minute normalized to protein content ± SD (n = 20 vehicle, n = 17 IPI), ****P < 0.0001, Mann–Whitney nonparametric test. (E) Representative image of MSK-PCa3 organoids treated with IPI or DMSO for 25 d (Left). Statistical analysis of the organoid sizes (Right). Diameters of organoids treated with IPI were compared with DMSO (n = 78 DMSO-treated, n = 95 IPI-treated), ****P < 0.0001, Student t test. Pixel magnification is indicated. (Scale bars, 200 pixels.)
FASN Protein Is Coexpressed with AR-FL and AR-V7 in Human mCRPC. To explore opportunities for therapy with IPI-9119 in the clinical mCRPC setting, we analyzed the expression of FASN, AR-FL, and AR-V7 proteins in tissue microarrays from 61 mCRPC patients. Six neuroendocrine cases were excluded from the analysis. Twenty-two of the remaining 55 cases were resistant to Enza and/or Abi. FASN was detected in 91% of metastatic sites, in significant association with AR-FL expression (87% of all metastases, P < 0.0001, Fisher’s exact test). AR-V7 positivity was observed in 39% of bone metastases, and 12% of visceral sites, and consistently coexpressed with FASN (Fig. 8 A and B and SI Appendix, Fig. S14). All but one patient had at least one metastatic site coexpressing FASN and AR-FL. In the Enza- and/or Abi-treated subset, FASN/AR-V7 double-positive metastases were found in 77% of cases. In addition, AR-V7 was detected in up to 25% of Enza/Abi treatment-naïve cases, always coexpressed with FASN (Fig. 8C). Individual FASN, AR-FL, and AR-V7 positivity and intensity for each case and metastatic site are described in detail in Dataset S1M.
FASN is coexpressed with AR-FL and AR-V7 in human mCRPCs. (A) Status of FASN, AR-FL, and AR-V7 in metastatic sites. Data are expressed as a percentage of either osseous or visceral metastases. (B) Representative images of FASN coexpression with AR-FL and AR-V7 (20×) in bone metastases: Bright field (a), FASN/AR-FL staining (b), FASN/AR-V7 staining in AR-V7–negative bone metastasis from a mCRPC Enza/Abi-naïve patient (c); bright field (d), FASN/AR-FL staining (e), FASN/AR-V7 staining in AR-V7–positive bone metastasis from a mCRPC Enza/Abi-naïve patient (f); bright field (g), FASN/AR-FL staining (h), FASN/AR-V7 staining in AR-V7-positive bone metastasis from a mCRPC Enza/Abi-treated patient (i). (C) Bar graph showing the percentage of patients with FASN/AR-FL and FASN/AR-V7 coexpression in all mCRPC patients analyzed or in the subset treated with Enza/Abi.
Taken together, these data provide the rationale for the use of FASN inhibitors in mCRPC overexpressing AR-V7, in both Enza- and/or Abi-resistant and -naïve patients.
DISCUSSION
Metabolic reprogramming contributes to treatment resistance. While normal prostatic cells mostly rely on dietary FAs, PCa cells increase the rate of de novo FA synthesis despite the presence of abundant circulating FAs. This metabolic phenotype is associated with PCa progression and androgen independence. De novo synthesized FAs contribute to new membrane synthesis, are a source of energy, promote oncogenic signaling, and contribute to drug resistance (13). Thus, lipogenesis represents a PCa-specific metabolic vulnerability that can be exploited therapeutically.
While FASN is an established therapeutic target, and FASN inhibitors had proved promising in the preclinical setting, off-target effects, poor solubility and pharmacokinetics, and untoward effects including weight loss have prevented their clinical development (13). Recently, newly released FASN inhibitors such as TVB-3166 (inhibitor of the β-ketoacyl-reductase domain) and Fasnall (a thiophenopyrimidine-based FASN inhibitor targeting the nucleotide binding pockets) showed encouraging results in patient-derived non-small-cell lung cancer xenografts and in the breast cancer murine MMTV-Neu model, respectively (22, 37). A modified, orally available version of TVB-3166, TVB-2640, is currently in phase-I clinical trials in patients with advanced solid malignancies (NCT02223247). Combined with paclitaxel, TVB-2640 proved beneficial in heavily pretreated breast cancer patients (38, 39). New trials are now investigating TVB-2640 in different stages of astrocytomas, breast, and colon cancers (NCT03032484, NCT03179904, and NCT02980029). However, no study has been conducted in mCRPC thus far.
We developed and characterized the selective, irreversible, and potent FASN inhibitor IPI-9119. We show that IPI-9119 significantly reduced cell growth and induced cell cycle arrest and apoptosis in PCa cells, while minimally affecting nontransformed cells. As expected, FASN inhibition markedly affected lipid homeostasis, including the synthesis and oxidation of FAs, as well as the metabolism of triglycerides and phospholipids. The accumulation of polyunsaturated FAs following IPI-9119 treatment suggests an attempt to compensate for the reduction in newly synthesized saturated FAs through increased uptake and utilization of exogenous FAs. Metabolic profiling uncovered an even broader metabolic rewiring, encompassing amino acid, carbohydrate, and TCA cycle pathways. Cholesterol levels and the expression of genes involved in steroidogenesis were increased following IPI-9119 treatment, as previously indicated (22). This effect can be ascribed, at least in part, to a redirection of the unused acetyl-CoA toward cholesterol synthesis. However, increased intratumoral steroidogenesis was not observed. The fate and biological implications of enhanced cholesterol synthesis warrant further investigation.
Interestingly, while the analysis of the metabolome/lipidome is consistent with the suppression of FASN activity, PCa cells attempt to compensate for de novo FA deficiency by up-regulating genes encoding enzymes and transcription factors involved in lipid synthesis, as previously found (22). Therefore, transcriptional changes should not be used as a readout of suppression of FASN activity.
FASN and FA synthesis are directly and indirectly regulated by the androgen/AR axis (19, 20). This paradigm holds true in mCRPC with reactivated AR signaling and FASN overexpression (15, 16). In addition, treatment with the SREBP inhibitor fatostatin and the cholesterol synthesis inhibitor simvastatin resulted in AR-FL protein level reduction (21, 40), suggesting that modulation of lipid metabolism may impact AR signaling.
While FASN inhibitors have shown efficacy in non-hormone-driven tumors as well as in AR-negative PCa cells (i.e., PC3 cells) (41), here we investigated the modulation of FA synthesis as an indirect, alternative approach to target the AR/AR-V7 pathway in the CRPC setting. We show that IPI-9119 results in the inhibition of AR-FL in both AD and AI cells harboring exclusively AR-FL and of AR-V7 in AI models driven by the variant (i.e., 22Rv1 and LNCaP-95). Both full-length and spliced AR protein were diminished in AD and AI PCa cells, while AR-FL reduction at the mRNA level was observed only in LNCaP cells. In fact, we show that ER stress induced by IPI-9119 results in the inhibition of protein translation and in AR/AR-V7 protein reduction. ER stress inhibitors partially rescue AR expression, suggesting that ER stress is involved in mediating the effect of FASN suppression on the AR pathway. The induction of ER stress had been previously associated with the down-regulation of the AR pathway through the reduction of AR protein synthesis as well as increased degradation (29, 30). Likewise, modulation of lipid metabolism and membrane lipid composition as well as inhibition of FASN activity have been all reported to trigger ER stress (26–28, 42). In AI cells driven by AR-V7, inhibition of de novo lipogenesis suppressed a gene signature derived from bone metastases with high levels of AR-V7 (33), as well as previously reported AR-V7–regulated gene signatures from in vitro models (32, 34). In addition, combination of IPI-9119 with Enza showed higher antitumor efficacy than monotherapy in an AR-V7–driven 22Rv1 cell line. Interestingly, IPI-9119 significantly reduced tumor growth in preclinical CRPC models resistant to Enza/Abi without overt side effects. Furthermore, FASN inhibition was also effective in an organoid model derived from human mCRPC. These findings have important therapeutic implications, particularly in the castration-resistant setting, in view of the finding that the overwhelming majority of human CRPC metastases we examined coexpressed FASN and AR-FL. Intriguingly, AR-V7 was positive in 39% of bone metastases, though only in a minority of visceral sites (12%). In both, FASN was consistently coexpressed with AR-V7. Therefore, FASN may be targeted in the majority of mCRPC, regardless of AR-V7 status (43). Inhibition of de novo lipogenesis could be proposed in association with Enza and/or Abi or taxanes, to delay/overcome resistance. Alternatively, FASN inhibition could be undertaken in tumors that are still AR-driven once resistance has emerged. Carefully designed clinical trials are required to establish the therapeutic timing, combinatorial regimens, and the population suitable for treatment with FASN inhibitors.
Finally, our data also indicate that protein expression of the AR-dependent gene c-MYC (44) as well as the c-MYC–regulated transcriptional signature are significantly reduced by FASN inactivation. These data are clinically relevant because c-MYC alterations occur in about 20% of mCRPC (45) and c-MYC transcript levels are increased in AR-V7–high bone metastases (33). Furthermore, overexpression of c-MYC protein was shown to induce androgen-independent growth (46). The potential involvement of c-MYC in alternative splicing and generation of AR-V7 has also been suggested (47). Our group had previously demonstrated that c-MYC overexpression is associated with pathway enrichment for FA metabolism in cell lines, murine models, and human prostate tumors. FASN expression was also increased in c-MYC–transformed cells, suggesting their dependency on de novo lipogenesis (48). In line with this, MYC-CaP cells were very sensitive to FASN inhibition with IPI-9119, suggesting a mutual regulation between FASN and c-MYC.
In conclusion, FASN inhibition rewires the CRPC metabolome and induces ER stress, resulting in the down-regulation of AR-FL/AR-V7 and CRPC cell growth inhibition. This study sets the stage and provides the rationale for clinical trials to test FASN inhibitors in mCRPC.
MATERIALS AND METHODS
Reagents.
IPI-9119 (US Patent nos. 8,546,432 and 9,346,769), developed by Infinity Pharmaceuticals, was resuspended in 100% DMSO for in vitro studies. For in vivo studies, IPI-9119 was resuspended in 20% 1-methyl-2-pyrrolidinone in 0.1 M sodium phosphate buffer, pH 8 (osmotic ALZET pump). All of the reagents used in the paper are described in SI Appendix.
Cell Culture.
Cells were cultured in standard conditions. Details are provided in SI Appendix. All cells were authenticated using short tandem repeat profiling performed by ATCC. A Mycoplasma test was performed bimonthly, using the colorimetric MycoAlert mycoplasma detection kit (Lonza). All cells used were tested negative.
Organoid Culture.
MSK-PCa3 organoids, kindly provided by Yu Chen, Memorial Sloan-Kettering Cancer Center, New York, were dissociated to single cells with trypsin for 15 min at 37 °C; 2 × 103 cells per well were mixed with 40 μL of growth factor reduced matrigel, seeded in a 24-well plate, and cultured in human organoid culture medium (36), in the presence of either FASN inhibitor or DMSO. Culture medium was refreshed every 3 d for 25 d. Organoids were imaged with a Zeiss Axio microscope, and their diameter was manually measured. Organoid samples were deidentified before use.
Cell Growth and Clonogenic Assays.
Measurement of cell growth and clonogenic assays were performed as previously described (49), with some modifications. Detailed information is provided in SI Appendix.
Cell Cycle and Apoptosis Studies.
Cell cycle and apoptosis analyses were performed as previously described (49, 50). Detailed information is provided in SI Appendix.
Immunoblotting.
Immunoblotting was performed using standard procedures. Detailed information is provided in SI Appendix.
Analysis of Protein Synthesis (SUnSET Assay).
SUnSET assay was performed to analyze protein synthesis, as per the manufacturer’s recommendations. Cells were treated with IPI-9119 for 3 and 6 d and then incubated with puromycin (1 µg/mL) for 30 min, before harvesting. Cells were washed with ice-cold PBS and lysed using 1% Nonidet P-40 buffer. An equal quantity of protein lysates was separated on SDS/PAGE and probed with antipuromycin antibody (kerafast).
Biochemical Assays and Oil Red O Staining.
FASN activity, de novo lipogenesis, FAO analysis, and Oil Red O staining were performed as previously described (48–51), with some modifications. Detailed information is provided in SI Appendix.
RNA Extraction and Real-Time qPCR.
RNA extraction and real-time qPCR were performed using standard procedures. Detailed information is provided in SI Appendix.
FASN and AR-FL/AR-V7 Genetic Manipulation.
Infections were performed as previously described (52) in biological triplicates. Details and construct information are provided in SI Appendix.
Luciferase Assay.
Luciferase assay was performed using the Dual Luciferase Assay Kit as described by the manufacturer. Detailed information is provided in SI Appendix.
RNA Sequencing.
Library preparation, sequencing, and data analysis were performed at the Center for Cancer Computational Biology. Detailed Information is provided in SI Appendix.
Metabolic Profiling.
Metabolic profiling was performed by the company Metabolon Inc., using their liquid chromatography tandem MS platforms. Information regarding sample preparation, quality assurance, quality control, metabolite identification/quantification, and data preprocessing is detailed in SI Appendix.
Lipidomics.
Lipids were extracted, dried under nitrogen steam, and store at −80 °C until further analysis. Lipidomic profiling was performed by the Mass Spectrometry, Metabolomics, and Proteomics core at Beth Israel Deaconess Medical Center using their high-resolution hybrid QExactive Plus Orbitrap mass spectrometer (Thermo Fisher Scientific), as described in ref. 53. Details are provided in SI Appendix.
Animal Studies.
Animal studies were performed according to ARRIVE guidelines and approved by the Dana-Farber Cancer Institute Institutional Care and Use Committee in accordance with the Guide for the Care and Use of Laboratory Animals (54). Details are provided in SI Appendix.
Human Studies and Image Analysis.
The human mCRPC specimens were obtained as part of the University of Washington (UW) Medical Center Prostate Cancer Donor Program, which was approved by the UW Institutional Review Board. Human studies were conducted according to Declaration of Helsinki principles. A written informed consent was received from participants before inclusion in the study. Description of the human samples and details on immunofluorescence staining, antibody dilution, and image analysis are provided in SI Appendix.
Statistical Analyses.
A detailed description of all of the statistical analyses is provided in SI Appendix. Differences were considered significant with P < 0.05 and, if multiple comparisons were done, FDR < 0.05. For GSEA and MSEA, an FDR threshold of 0.15 was used.
Data and Materials Availability.
RNA-seq data have been deposited in Gene Expression Omnibus (GEO) under accession no. GSE114016 (55). Raw and normalized metabolomics and lipidomics data are provided in Dataset S1 B and E–I.
ACKNOWLEDGMENTS
This work was supported by Department of Defense (DoD) IMPACT Grant PC160357 (to M.L., S.M.D., and S.R.P.), DoD synergistic Grant W81XWH1410405 (to M.L. and U.M.), NIH Grants R01-CA131945 and P50 CA90381, and the Prostate Cancer Foundation (PCF) (M.L.). G.Z. is a recipient of the DoD Idea Development Award for New Investigators (PC150263) and a Claudia Adams Barr Award in Innovative Cancer Research from the Dana-Farber Cancer Institute. The rapid autopsy material is the result of work supported by resources by the DoD (Award W81XWH-14-2-0183), the Pacific Northwest Prostate Cancer Specialized Programs of Research Excellence (SPORE) (Grant P50CA97186), and the Institute for Prostate Cancer Research. L.M.B. is supported by a Future Fellowship from the Australian Research Council (Fellowship FT130101004) and grant support from the Movember Foundation/PCF of Australia.
FOOTNOTES
Conflict of interest statement: A patent relative to the findings described in this study has been filed from the Dana-Farber Cancer Institute (52095-584P01US). J.T., A.C., K.M., V.J.P., J.A. and S.P. were former employees of Infinity Pharmaceuticals. J.L.K. is a current employee of Infinity Pharmaceuticals.
This article is a PNAS Direct Submission.
Data deposition: RNA sequencing data have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE114016).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1808834116/-/DCSupplemental.